Retrovirus
From Wikipedia, the free encyclopedia
| Retroviruses | ||||
|---|---|---|---|---|
| Virus classification | ||||
|
||||
| Genera | ||||
|
Subfamily: Orthoretrovirinae Subfamily: Spumaretrovirinae |
A retrovirus is a virus that does not travel and enter host cells with a DNA genome, but an RNA genome. The most common way the RNA genome is replicated is via the enzyme reverse transcriptase to make DNA out of its RNA genome. The DNA is then incorporated into the host's genome by an integrase enzyme. The virus thereafter replicates as part of the host cell's DNA. Retroviruses are enveloped viruses that belong to the viral family Retroviridae.
The virus itself stores its nucleic acid, in the form of a +mRNA (including the 5'cap and 3'PolyA inside the virion) genome and serves as a means of delivery of that genome into cells it targets as an obligate parasite, and constitutes the infection. Once in the host's cell, the RNA strands undergo reverse transcription in the cytosol and are integrated into the host's genome, at which point the retroviral DNA is referred to as a provirus. It is difficult to detect the virus until it has infected the host.
Contents |
[edit] Virion structure
Virions of retroviruses consist of enveloped particles about 100 nm in diameter. The virions also contain two identical single-stranded RNA molecules 7-10 kilobases (kb) in length. Although virions of different retroviruses do not have the same morphology or biology, all the virion components are very similar. [1]
The main virion components are:
- Envelope: composed of a lipid bilayer which is obtained from the host plasma membrane during budding process.
- RNA: consisted of a dimer RNA. It has a cap at 5' end and polyadenyle at 3' end. The RNA genome also has terminal noncoding regions which is important in replication and internal regions that encode virion proteins for gene expression. The 5' end includes four regions which are R, U5, PB and L. R region is a short repeated sequence at each end of the genome during the reverse transcription in order to ensure correct end-to-end transfer in growing chain. U5, on the other hand, is a short unique sequence between R and PB. PB consists of 18 bases complementary to 3' end of tRNA primer. L region is an untranslated leader region that gives signal for packaging of genome RNA. The 3' end includes 3 regions which are PP (polypurine), U3 and R. PP is primer for plus-strand DNA synthesis during reverse transcription. U3 is a sequence between PP and R which has signal that provirus can use in transcription. Lastly, R is the terminal repeated sequence at 3' end.
- Proteins: consisted of gag proteins, protease (PR), pol proteins and env proteins. Gag proteins are major components of the viral capsid which are about 2000-4000 copies per virion. Protease, on one hand, is expressed differently in different viruses. It functions in proteolytic cleavages during virion maturation to make mature gag and pol proteins. Pol proteins are responsible for synthesis of viral DNA and integration into host DNA after infection. Finally, env proteins play role in association and entry of virion into the host cell. [2]
[edit] Multiplication
When retroviruses have integrated their own genome into the germ line, their genome is passed on to a following generation. These endogenous retroviruses, contrasted with exogenous ones, now make up 5-8% of the human genome.[3] Most insertions have no known function and are often referred to as "junk DNA". However, many endogenous retroviruses play important roles in host biology, such as control of gene transcription, cell fusion during placental development in the course of the germination of an embryo, and resistance to exogenous retroviral infection. Endogenous retroviruses have also received special attention in the research of immunology-related pathologies, such as autoimmune diseases like multiple sclerosis, although endogenous retroviruses have not yet been proven to play any causal role in this class of disease. The role of endogenous retroviruses in human gene evolution is explored in a 2005 peer-reviewed article.[4]
While transcription was classically thought to only occur from DNA to RNA, reverse transcriptase transcribes RNA into DNA. The term "retro" in retrovirus refers to this reversal (making DNA from RNA) of the central dogma of molecular biology. Reverse transcriptase activity outside of retroviruses has been found in almost all eukaryotes, enabling the generation and insertion of new copies of retrotransposons into the host genome. These inserts are transcribed by enzymes of the host into new RNA molecules which enter the cytosol. Next, some of these RNA molecules are translated into viral proteins. For example, the gag gene is translated into molecules of the capsid protein, the pol gene is transcribed into molecules of reverse transcriptase, and the env gene is translated into molecules of the envelope protein. It is important to note that a retrovirus must "bring" its own reverse transcriptase in its capsid, otherwise it is unable to utilize the enzymes of the infected cell to carry out the task, due to the unusual nature of producing DNA from RNA.
Industrial drugs that are designed as protease and reverse transcriptase inhibitors can quickly be proved ineffective because the gene sequences that code for the protease and the reverse transcriptase can undergo many substitutions. These substitutions of nitrogenous bases, which make up the DNA strand, can make either the protease or the reverse transcriptase difficult to attack. The amino acid substitution enables the enzymes to evade the drug regiments because mutations in the gene sequences can cause physical or chemical change which makes them harder to detect by the drug. When the drugs that are supposed to attack enzymes, such as protease, are designed, the manufacturers target specific sites on the enzyme. One way to attack these targets can be through hydrolysis of molecular bonds, which means that the drug will add molecules of H2O (water) to specific bonds. By adding molecules of water at a site on the virus, the drug breaks the previous bonds that were linked to each other. If several of these breaks occur, the result can lead to lysis, the death of the virus. [5]
Because reverse transcription lacks the usual proofreading of DNA replication, a retrovirus mutates very often. This enables the virus to grow resistant to antiviral pharmaceuticals quickly, and impedes the development of effective vaccines and inhibitors for the retrovirus.[6]
[edit] Genes
Retrovirus genomes commonly contain these three open reading frames that encode for proteins that can be found in the mature virus:
- group-specific antigen (gag) codes for core and structural proteins of the virus;
- polymerase (pol) codes for reverse transcriptase, protease and integrase; and,
- envelope (env) codes for the retroviral coat proteins.
[edit] Provirus
This DNA can be incorporated into host genome as a provirus that can be passed on to progeny cells. In this way some retroviruses can convert normal cells into cancer cells. Some provirus remains latent in the cell for a long period of time before it is activated by the change in cell environment.
[edit] Development
Studies of retroviruses led to the first demonstrated synthesis of DNA from RNA templates, a fundamental mode for transferring genetic material that occurs in both eukaryotes and prokaryotes. It has been speculated that the RNA to DNA transcription processes used by retroviruses may have first caused DNA to be used as genetic material. In this model, the RNA world hypothesis, cellular organisms adopted the more chemically stable DNA when retroviruses evolved to create DNA from the RNA templates.
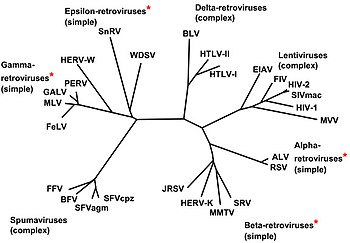
[edit] Classification
[edit] Exogenous
The following genera are included here:
- Genus Alpharetrovirus; type species: Avian leukosis virus
- Genus Betaretrovirus; type species: Mouse mammary tumour virus
- Genus Gammaretrovirus; type species: Murine leukemia virus; others include Feline leukemia virus
- Genus Deltaretrovirus; type species: Bovine leukemia virus; others include the cancer-causing Human T-lymphotropic virus
- Genus Epsilonretrovirus; type species: Walleye dermal sarcoma virus
- Genus Lentivirus; type species: Human immunodeficiency virus 1; others include Simian, Feline immunodeficiency viruses
- Genus Spumavirus; type species: Chimpanzee foamy virus
These were previously divided into three subfamilies (Oncovirinae, Lentivirinae, and Spumavirinae), but with current knowledge of retroviruses, this is no longer appropriate. (The term oncovirus is still commonly used, though.)
[edit] Endogenous
Endogenous retroviruses are not formally included in this classification system, and are broadly classified into three classes, on the basis of relatedness to exogenous genera:
- Class I are most similar to the gammaretroviruses
- Class II are most similar to the betaretroviruses and alpharetroviruses
- Class III are most similar to the spumaviruses
[edit] Treatment
Antiretroviral drugs are medications for the treatment of infection by retroviruses, primarily HIV. Different classes of antiretroviral drugs act at different stages of the HIV life cycle. Combination of several (typically three or four) antiretroviral drugs is known as highly active anti-retroviral therapy (HAART).
[edit] Treatment of animal infections
Feline Leukemia Virus and Feline immunodeficiency virus infections are treated with biologics, including Lymphocyte T-Cell Immune Modulator[7]
[edit] Genetic barrier
The genetic barrier is loosely defined as the difficulty for retroviruses to evade antiretroviral drugs by mutating into resistant types. [8]
[edit] References
- ^ Jhon M. Coffin (1992). "Structure and Classification of Retroviruses". in Jay A. Levy. The Retroviridae (1st ed.). New York: Plenum Press. pp. 20. ISBN 0-306-44074-1.
- ^ Jhon M. Coffin (1992). "Structure and Classification of Retroviruses". in Jay A. Levy. The Retroviridae (1st ed.). New York: Plenum Press. pp. 26-34. ISBN 0-306-44074-1.
- ^ Robert Belshaw; Pereira V; Katzourakis A; Talbot G; Paces J; Burt A; Tristem M. (April 2004). "Long-term reinfection of the human genome by endogenous retroviruses". Proc Natl Acad Sci U S A 101 (14): 4894–99. doi:. PMID 15044706. http://www.pubmedcentral.com/articlerender.fcgi?artid=387345.
- ^ Medstrand P, van de Lagemaat L, Dunn C, Landry J, Svenback D, Mager D (2005). "Impact of transposable elements on the evolution of mammalian gene regulation". Cytogenet Genome Res 110 (1-4): 342–52. doi:. PMID 16093686.
- ^ {{|Teklemariam, Ephrem |http://mason.gmu.edu/~eteklema/Researchfinal.html |HIV drug resistance and its impact on public health.|joRes
- ^ Svarovskaia ES; Cheslock SR; Zhang WH; Hu WS; Pathak VK. (January 2003). "Retroviral mutation rates and reverse transcriptase fidelity.". Front Biosci. 8: 117–134. doi:. http://www.ncbi.nlm.nih.gov/pubmed/12456349.
- ^ [1]
- ^ Computing the genetic barrier N. Beerenwinkel, T. Sing, M. Daumer, R. Kaiser, T. Lengauer.
[edit] External links
- Retrovirus Animation (Flash Required)
- Retrovirology Scientific journal
- Retroviruses at rcn.com
- NCBI retrovirus book online
- Annals of Science: Darwin’s Surprise, New Yorker Dec 3 2007
|
||||||||||||||
|
|||||||||||||||||||||||||||||||