Protein folding
From Wikipedia, the free encyclopedia
Protein folding is the physical process by which a polypeptide folds into its characteristic and functional three-dimensional structure.[1]
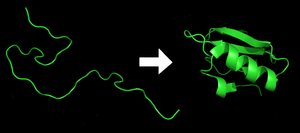
Each protein begins as a polypeptide, translated from a sequence of mRNA as a linear chain of amino acids. This polypeptide lacks any developed three-dimensional structure (the left hand side of the neighboring figure). However each amino acid in the chain can be thought of having certain 'gross' chemical features. These may be hydrophobic, hydrophilic, or electrically charged, for example. These interact with each other and their surroundings in the cell to produce a well-defined, three dimensional shape, the folded protein (the right hand side of the figure), known as the native state. The resulting three-dimensional structure is determined by the sequence of the amino acids.[2] The mechanism of protein folding is not completely understood.
Experimentally determining the three dimensional structure of a protein is often very difficult and expensive. However the sequence of that protein is often known. Therefore scientists have tried to use different biophysical techniques to computationally fold a protein, that is, to predict the structure of the complete protein from the sequence of the protein.
For many proteins the correct three dimensional structure is essential to function.[3] Failure to fold into the intended shape usually produces inactive proteins with different properties (details found under prion). Several neurodegenerative and other diseases are believed to result from the accumulation of misfolded (incorrectly folded) proteins.[4]
Contents |
[edit] Known facts about the process
[edit] The relationship between folding and amino acid sequence
The amino-acid sequence (or primary structure) of a protein predisposes it towards its native conformation or conformations. It will fold spontaneously during or after synthesis. While these macromolecules may be regarded as "folding themselves", the mechanism depends equally on the characteristics of the cytosol, including the nature of the primary solvent (water or lipid), macromolecular crowding,[5] the concentration of salts, the temperature, and molecular chaperones.
Most folded proteins have a hydrophobic core in which side chain packing stabilizes the folded state, and charged or polar side chains on the solvent-exposed surface where they interact with surrounding water molecules. It is generally accepted that minimizing the number of hydrophobic side-chains exposed to water is the principal driving force behind the folding process,[6] although a recent theory has been proposed which reassesses the contributions made by hydrogen bonding.[7]The strengths of hydrogen bonds in a protein vary, i.e. they are dependent on their microenvironment, thus H-bonds enveloped in a hydrophobic core contribute more than H-bonds exposed to the aqueous environment to the stability of the native state.[8]
The process of folding in vivo often begins co-translationally, so that the N-terminus of the protein begins to fold while the C-terminal portion of the protein is still being synthesized by the ribosome. Specialized proteins called chaperones assist in the folding of other proteins.[9] A well studied example is the bacterial GroEL system, which assists in the folding of globular proteins. In eukaryotic organisms chaperones are known as heat shock proteins. Although most globular proteins are able to assume their native state unassisted, chaperone-assisted folding is often necessary in the crowded intracellular environment to prevent aggregation; chaperones are also used to prevent misfolding and aggregation which may occur as a consequence of exposure to heat or other changes in the cellular environment.
For the most part, scientists have been able to study many identical molecules folding together en masse. At the coarsest level, it appears that in transitioning to the native state, a given amino acid sequence takes on roughly the same route and proceeds through roughly the same intermediates and transition states. Often folding involves first the establishment of regular secondary and supersecondary structures, particularly alpha helices and beta sheets, and afterwards tertiary structure. Formation of quaternary structure usually involves the "assembly" or "coassembly" of subunits that have already folded. The regular alpha helix and beta sheet structures fold rapidly because they are stabilized by intramolecular hydrogen bonds, as was first characterized by Linus Pauling. Protein folding may involve covalent bonding in the form of disulfide bridges formed between two cysteine residues or the formation of metal clusters. Shortly before settling into their more energetically favourable native conformation, molecules may pass through an intermediate "molten globule" state.
The essential fact of folding, however, remains that the amino acid sequence of each protein contains the information that specifies both the native structure and the pathway to attain that state. This is not to say that nearly identical amino acid sequences always fold similarly.[10] Conformations differ based on environmental factors as well; similar proteins fold differently based on where they are found. Folding is a spontaneous process independent of energy inputs from nucleoside triphosphates. The passage of the folded state is mainly guided by hydrophobic interactions, formation of intramolecular hydrogen bonds, and van der Waals forces, and it is opposed by conformational entropy.
[edit] Disruption of the native state
In certain solutions and under some conditions proteins will not fold into their biochemically functional forms. Temperatures above (and sometimes those below) the range that cells tend to live in will cause thermally unstable proteins to unfold or "denature" (this is why boiling makes an egg white turn opaque). High concentrations of solutes, extremes of pH, mechanical forces, and the presence of chemical denaturants can do the same. A fully denatured protein lacks both tertiary and secondary structure, and exists as a so-called random coil. Under certain conditions some proteins can refold; however, in many cases denaturation is irreversible.[11] Cells sometimes protect their proteins against the denaturing influence of heat with enzymes known as chaperones or heat shock proteins, which assist other proteins both in folding and in remaining folded. Some proteins never fold in cells at all except with the assistance of chaperone molecules, which either isolate individual proteins so that their folding is not interrupted by interactions with other proteins or help to unfold misfolded proteins, giving them a second chance to refold properly. This function is crucial to prevent the risk of precipitation into insoluble amorphous aggregates.
[edit] Incorrect protein folding and neurodegenerative disease
Aggregated proteins are associated with prion-related illnesses such as Creutzfeldt-Jakob disease, bovine spongiform encephalopathy (mad cow disease), amyloid-related illnesses such as Alzheimer's Disease and familial amyloid cardiomyopathy or polyneuropathy, as well as intracytoplasmic aggregation diseases such as Huntington's and Parkinson's disease. These age onset degenerative diseases are associated with the multimerization of misfolded proteins into insoluble, extracellular aggregates and/or intracellular inclusions including cross-beta sheet amyloid fibrils; it is not clear whether the aggregates are the cause or merely a reflection of the loss of protein homeostasis, the balance between synthesis, folding, aggregation and protein turnover. Misfolding and excessive degradation instead of folding and function leads to a number of proteopathy diseases such as antitrypsin-associated Emphysema, cystic fibrosis and the lysosomal storage diseases, where loss of function is the origin of the disorder. While protein replacement therapy has historically been used to correct the latter disorders, an emerging approach is to use pharmaceutical chaperones to fold mutated proteins to render them functional. Christopher M. Dobson, Jeffery W. Kelly, Dennis Selkoe, Stanley Prusiner, Peter T. Lansbury, William E. Balch, Richard I. Morimoto, Susan L. Lindquist and Byron C. Caughey have all contributed to this emerging understanding of diseases that are some of the most menacing of our era.
[edit] Kinetics and the Levinthal Paradox
The entire duration of the folding process varies dramatically depending on the protein of interest. The slowest folding proteins require many minutes or hours to fold, primarily due to proline isomerizations or wrong disulfide bond formations, and must pass through a number of intermediate states, like checkpoints, before the process is complete.[12] On the other hand, very small single-domain proteins with lengths of up to a hundred amino acids typically fold in a single step.[13] Time scales of milliseconds are the norm and the very fastest known protein folding reactions are complete within a few microseconds.[14]
The Levinthal paradox[15] observes that if a protein were to fold by sequentially sampling all possible conformations, it would take an astronomical amount of time to do so, even if the conformations were sampled at a rapid rate (on the nanosecond or picosecond scale). Based upon the observation that proteins fold much faster than this, Levinthal then proposed that a random conformational search does not occur in folding, and the protein must, therefore, fold by a directed process.
[edit] Techniques for studying protein folding
[edit] Circular Dichroism
Circular dichroism is one of the most general and basic tools to study protein folding. Circular dichroism spectroscopy measures the absorption of circularly polarized light. In proteins, structures such as alpha helicies and beta sheets are chiral, and thus absorb such light. The absorption of this light acts as a marker of the degree of foldedness of the protein ensemble. This technique can be used to measure equilibrium unfolding of the protein by measuring the change in this absorption as a function of denaturant concentration or temperature. A denaturant melt measures the free energy of unfolding as well as the protein's m value, or denaturant dependence. A temperature melt measures the melting temperature (Tm) of the protein. This type of spectroscopy can also be combined with fast-mixing devices, such as stopped flow, to measure protein folding kinetics and to generate chevron plots.
[edit] Dual Polarization Interferometry
Dual polarisation interferometry (DPI) has emerged over the past decade as an analytical technique that can probe protein layers adsorbed to the surface of a waveguide by using the evanescent wave of a laser beam confined to the waveguide.
DPI focuses laser light into two waveguides, one, the "sensing" waveguide, with an exposed surface and one to create a reference beam. A two-dimensional interference pattern is formed in the far field by combining the light passing through the two waveguides. The DPI technique uses two polarisation of the laser to excite two polarisation modes of the waveguides. Measurement of the interferogram for both polarisations allows both the refractive index (protein density or fold) and the size (conformation) of the adsorbed layer to be calculated. Real time measurements of biochemistry take place in a flow-through system. These measurements can be used to infer structural information about the molecular interactions at sub atomic resolution and is typically used to characterise biochemical interactions.
The latest versions of Dual Polarisation Interferometers also have the capability to quantify the order and disruption in birefringent thin films. This has been used, for example, to study the formation of lipid bilayers and their interaction with membrane proteins.
[edit] Modern studies of folding with high time resolution
The study of protein folding has been greatly advanced in recent years by the development of fast, time-resolved techniques. These are experimental methods for rapidly triggering the folding of a sample of unfolded protein, and then observing the resulting dynamics. Fast techniques in widespread use include ultrafast mixing of solutions, photochemical methods, and laser temperature jump spectroscopy. Among the many scientists who have contributed to the development of these techniques are Heinrich Roder, Harry Gray, Martin Gruebele, Brian Dyer, William Eaton, Sheena Radford, Chris Dobson, Sir Alan R. Fersht and Bengt Nölting.
[edit] Energy landscape theory of protein folding
The protein folding phenomenon was largely an experimental endeavor until the formulation of energy landscape theory by Joseph Bryngelson and Peter Wolynes in the late 1980s and early 1990s. This approach introduced the principle of minimal frustration, which asserts that evolution has selected the amino acid sequences of natural proteins so that interactions between side chains largely favor the molecule's acquisition of the folded state. Interactions that do not favor folding are selected against, although some residual frustration is expected to exist. A consequence of these evolutionarily selected sequences is that proteins are generally thought to have globally "funneled energy landscapes" (coined by José Onuchic) that are largely directed towards the native state. This "folding funnel" landscape allows the protein to fold to the native state through any of a large number of pathways and intermediates, rather than being restricted to a single mechanism. The theory is supported by both computational simulations of model proteins and numerous experimental studies, and it has been used to improve methods for protein structure prediction and design.
[edit] Computational prediction of protein tertiary structure
De novo or ab initio techniques for computational protein structure prediction is related to, but strictly distinct from, studies involving protein folding. Molecular Dynamics (MD) is an important tool for studying protein folding and dynamics in silico. Because of computational cost, ab initio MD folding simulations with explicit water are limited to peptides and very small proteins. MD simulations of larger proteins remain restricted to dynamics of the experimental structure or its high-temperature unfolding. In order to simulate long time folding processes (beyond about 1 microsecond), like folding of small-size proteins (about 50 residues) or larger, some approximations or simplifications in protein models need to be introduced. An approach using reduced protein representation (pseudo-atoms representing groups of atoms are defined) and statistical potential is not only useful in protein structure prediction, but is also capable of reproducing the folding pathways.[16]
Because of the many possible ways of folding, there can be many possible structures. A peptide consisting of just five amino acids can fold into over 100 billion possible structures.[17]
[edit] Techniques for determination of protein structure
The determination of the folded structure of a protein is a lengthy and complicated process, involving methods like X-ray crystallography and NMR. One of the major areas of interest is the prediction of native structure from amino-acid sequences alone using bioinformatics and computational simulation methods.
There are distributed computing projects which use idle CPU time of personal computers to solve problems such as protein folding or prediction of protein structure. People can run these programs on their computer or PlayStation 3 to support them. See links below (for example Folding@Home) to get information about how to participate in these projects.
[edit] See also
- Protein structure prediction
- Folding (chemistry)
- Anfinsen's dogma
- Levinthal paradox
- Denaturation (biochemistry)
- Protein design
- Chevron plot
- Denaturation midpoint
- Equilibrium unfolding
- Folding@Home
- Rosetta@Home
- Downhill folding
- Foldit computer game
- Software for molecular mechanics modeling
[edit] References
- ^ Alberts, Bruce; Alexander Johnson, Julian Lewis, Martin Raff, Keith Roberts, and Peter Walters (2002). "The Shape and Structure of Proteins". Molecular Biology of the Cell; Fourth Edition. New York and London: Garland Science. ISBN 0-8153-3218-1.
- ^ Anfinsen C (1972). "The formation and stabilization of protein structure". Biochem. J. 128 (4): 737–49. PMID 4565129.
- ^ Jeremy M. Berg, John L. Tymoczko, Lubert Stryer; Web content by Neil D. Clarke (2002). "3. Protein Structure and Function". Biochemistry. San Francisco: W.H. Freeman. ISBN 0-7167-4684-0.
- ^ "Science of Folding@Home". July 18, 2005. http://folding.stanford.edu/science.html. Retrieved on 2007-04-22.
- ^ van den Berg B, Wain R, Dobson CM, Ellis RJ (August 2000). "Macromolecular crowding perturbs protein refolding kinetics: implications for folding inside the cell". Embo J. 19 (15): 3870–5. doi:. PMID 10921869.
- ^ Pace C, Shirley B, McNutt M, Gajiwala K (1996). "Forces contributing to the conformational stability of proteins". Faseb J. 10 (1): 75–83. PMID 8566551. http://www.fasebj.org/cgi/reprint/10/1/75.
- ^ Rose G, Fleming P, Banavar J, Maritan A (2006). "A backbone-based theory of protein folding". Proc. Natl. Acad. Sci. U.S.A. 103 (45): 16623–33. doi:. PMID 17075053.
- ^ Deechongkit S, Nguyen H, Dawson PE, Gruebele M, Kelly JW (2004). "Context Dependent Contributions of Backbone H-Bonding to β-Sheet Folding Energetics". Nature 403 (45): 101–105. doi:. PMID 17075053.
- ^ Lee S, Tsai F (2005). "Molecular chaperones in protein quality control". J. Biochem. Mol. Biol. 38 (3): 259–65. PMID 15943899. http://www.jbmb.or.kr/fulltext/jbmb/view.php?vol=38&page=259.
- ^ Alexander PA, He Y, Chen Y, Orban J, Bryan PN. (2007). "The design and characterization of two proteins with 88% sequence identity but different structure and function". Proc Natl Acad Sci U S A. 104 (29): 11963–8. doi:. PMID 17609385. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pubmed&pubmedid=17609385.
- ^ Shortle D (1996). "The denatured state (the other half of the folding equation) and its role in protein stability". Faseb J. 10 (1): 27–34. PMID 8566543. http://www.fasebj.org/cgi/reprint/10/1/27.
- ^ P.S. Kim & R.L. Baldwin (1990). "Intermediates in the folding reactions of small proteins". Annu. Rev. Biochem. 59: 631–660. doi:.
- ^ S.E. Jackson (August 1998). "How do small single-domain proteins fold?" ([dead link] – Scholar search). Fold. Des. 3: R81–R91. doi:. ISSN 1359-0278. http://biomednet.com/elecref/13590278003R0081.
- ^ J. Kubelka, et al. (2004). "The protein folding "speed limit"". Curr. Opin. Struct. Biol. 14: 76–88. doi:.
- ^ C. Levinthal (1968). "Are there pathways for protein folding?". J. Chim. Phys. 65: 44–45.
- ^ Kmiecik S and Kolinski A (2007). "Characterization of protein-folding pathways by reduced-space modeling". Proc. Natl. Acad. Sci. U.S.A. 104 (30): 12330–12335. doi:. PMID 17636132.
- ^ Steven Schultz (1999). "Math helps explain protein folding". Princeton Weekly Bulletin 89 (3). http://www.princeton.edu/pr/pwb/99/0927/math.shtml.
[edit] External links
|
|||||||||||||||||||||||
|
|||||||||||